Hardtech is becoming (somewhat) easier: the role of simulations
Advances in simulations are reducing the cost of every iteration and therefore, eventual risk
In the previous posts, I talked about what kind of technologies make up hardtech and why investing in these technologies can seem difficult. In this post (and two following) I will talk about why hardtech is becoming somewhat easier to build and commercialise. Let’s dive right in.
Nowhere else is the difference between hardtech and all other tech more apparent than in the cost of a single iteration.
When you are building things in hardtech, each choice you make regarding the design of an iteration is a decision that cuts through multiple disciplines of sciences. To complicate things further, each choice that you make will undoubtedly have huge implications when you move towards mass-scale manufacturing.
You can’t just move fast and break things. If you do move fast, you will break a lot more things down the line. And it is never worth it. Hardtech development focuses on retiring risks, serially and diligently. The problem, however, is that every risk retirement exercise is an iteration in itself. Every single iteration in hardtech has traditionally been expensive. This is compounded by the fact that many failed expensive experiments are needed to arrive upon the first successful iteration.
The cost of iteration in hardtech, hence, is one of the biggest hurdles that inhibits migration of ideas from concept to product. Cost, in this context, arises from a lack of clarity. Specifically, two questions regarding an iteration could never be convincingly answered before.
Question #1:
Which of your designs should you prototype?
Given a set of parameters of operation, how can you “forecast” what is most likely to work and compare it to other versions that are less likely to? How can you choose which MVP to build based on “forecasted” outcome?
Question #2:
Can you make your prototype to your desired specifications?
How does your tangible prototype differ from the modelled specifications used to forecast performance? What effect does it have on your chances of success?
In the face of overwhelming fuzziness on these two counts, it is not difficult to imagine that resources get misallocated and priorities get misplaced. Once you are locked in to a wrong choice, course correcting becomes expensive (if at all possible).
This is where most hardtech ideas die —in search of the successful iteration that can scale.
This is now changing. And it is making hardtech, somewhat easier.
In this post, we will talk about how we are getting really good at answering question #1. In the next, we will tackle question #2.
Learning to predict the future: simulations
“Look before you leap” is an ancient proverb. For hardtech development, it is also sound and critical advice. Simulating the behaviour of a system is the closest a hardtech developer can come to “looking” before making the proverbial leap. This is not a novel concept in any way. Engineers and scientists have been simulating behaviours of systems they are interested in as far back as World War II. At any given point in our technological history, however, our ability to simulate systems has always been throttled by the computational power commonly available.
Real-life systems are complex. Modelling their behaviour is a computationally demanding problem. Scientists have been developing computational tools and programming techniques that matched specific problems and methods in their respective fields: reducing a problem to simpler components, or framing it in terms of phenomenon that are unquestionably true.
When there is not enough computational power available to throw at the problem, we resort to approximations to simplify the computation. The deviation of an approximation from the real “state” of an object is the primary cause of deviation between modelled and actual behavior. Hence, the biggest challenge of previous years has been to make the right approximations, “tuning” our models to make more accurate predictions within the constraints of available computational power.
This choice was simultaneously the solution and the problem.
Running simulations wasn’t cheap. Even after allocating the resources for computing infrastructure and expensive manpower, the predictions (for the above described problems) didn’t seem to help as they were highly divergent from actual observations. Success, if any, was random and rarely, if ever, replicable. As a result, modelling earned a bad reputation for being mere speculation.

The effect of increasing computational power on our ability to predict:
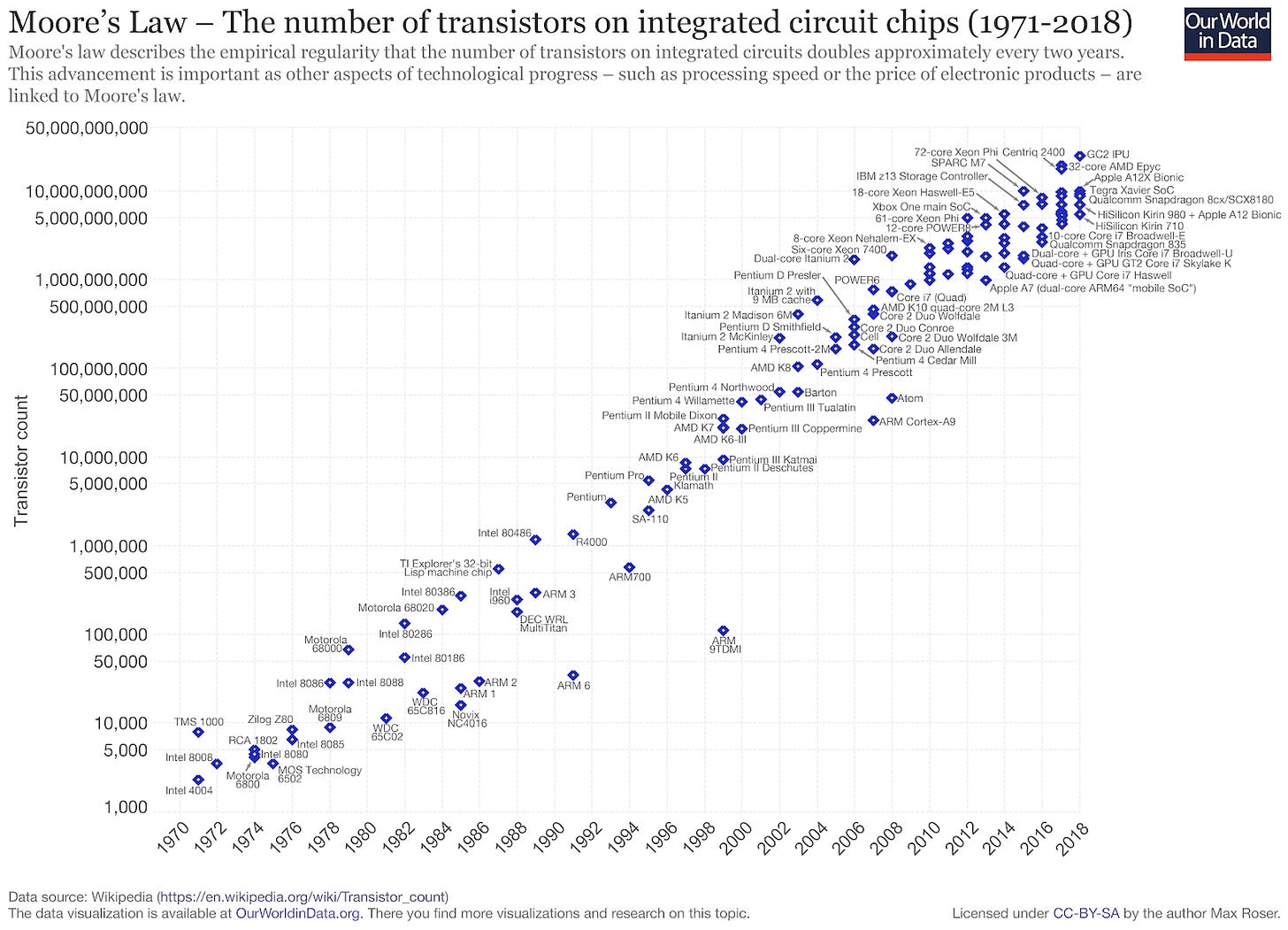
One of the most used graphs in technology is the one that shows Moore’s Law in action: the number of transistors on a chip doubles every two years. Additionally, we pay less for each transistor. In 2000, Intel’s Pentium 4 had 50M transistors on a chip. The Apple M1 chip powering the latest 2020 Macbook has 16B. Nvidia’s latest GA100 GPU has 54B.
When computational power is no longer a limiting factor, we start seeing a wonderful ecosystem level effect. Sure, the video games we love go from looking like coloured pixels to being indistinguishable from movies. More importantly though, we need to make lesser and lesser approximations. The need to oversimplify our models to make them computationally tractable is drastically reduced (not eliminated). In turn, our predictions start moving towards actually observed behaviour. When this happens often enough, every stakeholder stops thinking that computer modelling is mere speculation and starts believing that this can become a critical force multiplier. As more research gets funded, more knowledge gets created, algorithms get better, more talent is generated and they take this skill far and wide.

Out in the real world, proof of progress:
Let’s start looking at the effect of computational modelling by examining a long-standing problem in life sciences: protein folding.
Protein folding is a problem that has been attacked through computer simulations since about 1990. Proteins are strings of individual amino acids. Proteins “fold” into particular shapes which gives them their particular function. Over the past five decades, researchers have been able to determine shapes of proteins in labs using experimental techniques like cryo-electron microscopy, nuclear magnetic resonance and X-ray crystallography, but each method depends on a lot of trial and error, which can take years of work, and cost tens or hundreds of thousands of dollars per protein structure. Obviously, this is a critical rate-limiting step in biological discovery processes. The question: given a string of amino acids, can we predict what its shape would be and hence, by extension, what can it be expected to do? (For more details, read section “proteins are the building blocks of life” in the first issue of this newsletter)
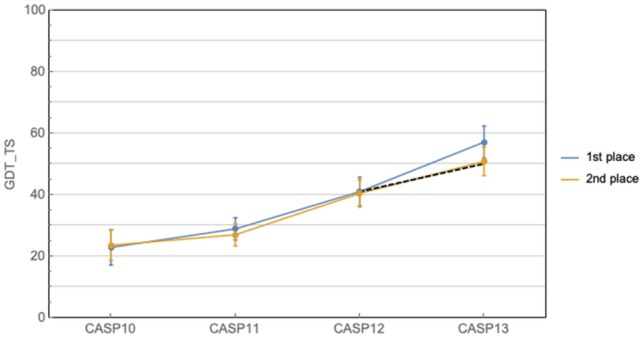
Alphafold, created by Deepmind blew the field of protein structure prediction wide open in 2018 when they could accurately predict the structure of 24 out of 43 protein domains having known only the sequence of amino acids. Admittedly, other groups had been making progress and getting better over the years anyway. Yet the application of deep learning backed by an unknown allocation of GPU resources was enough to allow Alphafold to pull away significantly from the course of expected technological year-on-year progress. Think of the Y-axis as how close the prediction is to the real shape of the protein. The difference between the 1st and 2nd placed algorithms at this specific competition called CASP has probably never been as large as it was in 2018.
In 2020, Alphafold took this further. Based on its performance, Alphafold 2 was recognised as a possible solution to the protein folding problem. John Moult, a structural biologist at the University of Maryland, Shady Grove, and co-founder of the competition, Critical Assessment of Protein Structure Prediction (CASP) exclaimed, “this is a 50 year old problem. I never thought I’d see this in my lifetime.”
You can also read AlphaFold’s success in predicting protein structure as the triumph of deep learning over more mathematical approaches grounded in biophysics. You will not be wrong. But it only supports my claim: none of this is possible without having the computational power that we have today.
It is important to note that these are not merely intellectual endeavours anymore. Computationally designed proteins are now maturing as real-world applications. In 2011, a team consisting of undergraduates of University of Washington modified an enzyme produced by a bacteria and used it to digest the gluten protein which causes celiac disease in sensitive populations. Over the years, the team used computational design to modify the enzyme, eventually landing a deal with Takeda who would invest $35M in 2018 for continued research, in exchange of rights to acquire the company if the research was borne out by clinical experimentation. In 2020, Takeda exercised that option, buying the startup, named PvP Biologics for $330M.
This ability doesn’t just supercharge life sciences.
SpaceX has been vocal about the use of simulations and computer-aided design software in their design process for a long time. Over and above design as such, the bane of engine development is the inability to “look inside” the combustion process itself — what happens when fuel and oxidizers introduced through distinct streams meet. Obviously, designing engines is an extremely expensive iteration process and nothing can possibly be more useful than to weed out bad hypotheses with simulations rather than needing to actually learn from a manufactured prototype’s failure. For SpaceX specifically, simulations were critical: failure to capture all relevant parameters in a model or over-approximating any of them would lead to a vicious and catastrophic interplay of non linear physical phenomenon, the result of which is dangerous. You can have an engine that explodes or one that shakes so violently that you can’t mount a payload on it. There are an incredible number of parameters that can be tweaked and the ability to model outcomes with each of those knobs slightly tweaked, continuously seeking to refine the design of the best engine and identifying its corresponding best operating parameters is a humongous computational problem. Without acceleration from GPUs, the simplest simulations in these domains would take months to run on thousands of cores. Now, SpaceX says they do this everyday.
The AWS effect: democratisation of simulations
So now that we have talked about how computational modelling of complex phenomena can lead to game-changing advantages for hardtech, we can start discussing access to such computational power. High-performance computing infrastructure is prohibitively expensive. A HPC cluster which can run simulations of meaningful impact will rarely, if ever, cost anything below a few hundred thousands of dollars. Apart from this upfront cost, there are several other maintenance costs that quickly add up. Hence, the pool of engineers/scientists who have access to these clusters swiftly become limited to those with affiliation to research institutions/labs funded by governments or large corporations. As a result, without such affiliation, hardtech founders have an entry barrier to deal with.
Enter: the HPC cloud
The ability to pay for compute and storage used has democratised access to HPC infrastructure. Cloud computing has made it easy for early teams to run valuable experiments, generate proof-of-concept level data and convert some believers before needing to invest any significant amount of resources into building on-premise infrastructure. Now, you would need tens of thousands (or lesser) to generate the same level of critical data.
How does this change the way that hardtech companies can be started? Let’s look at Reverie Labs. Reverie Labs is a company that is developing next generation kinase inhibitors which target brain tumors. Typically, drug developers will spend $ 100s of M to identify the first few lead drug candidates that will proceed through the rest of the development pipeline. Reverie uses deep learning techniques to screen candidate drugs, optimise them and suggest possible changes. They are now partnered with the biopharma giant Roche and Genentech.
The most important part of this journey is the start. The founders started a drug development company from a NY city apartment with 3 laptops, no lab of their own and no licensed assets or IP!
A tidal wave of simulation-aided hardtech companies is gathering on the horizon. Watch out for this wave to hit shore in the next few years.
In the next part, I will talk about how precision in manufacturing is allowing more simulations to be fabricated into prototypes with lesser chances of failure.
If you have any feedback, I would love to hear from you.